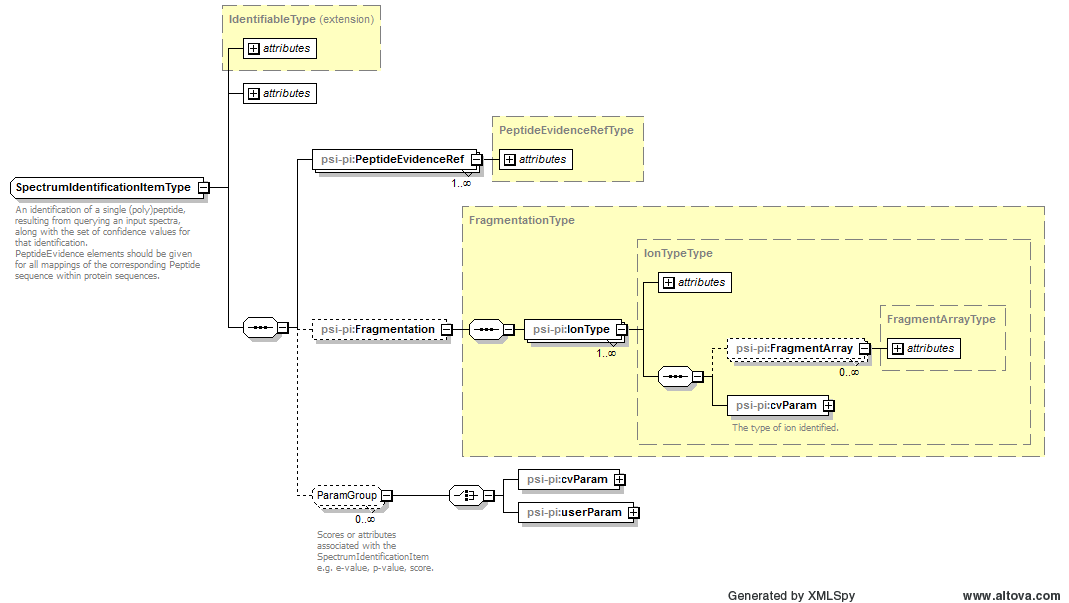
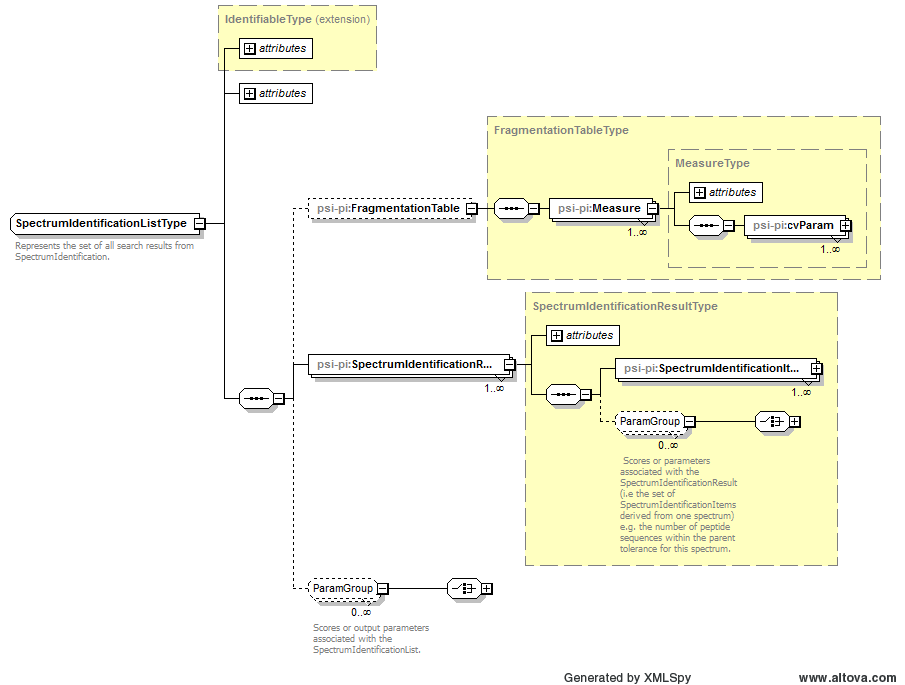
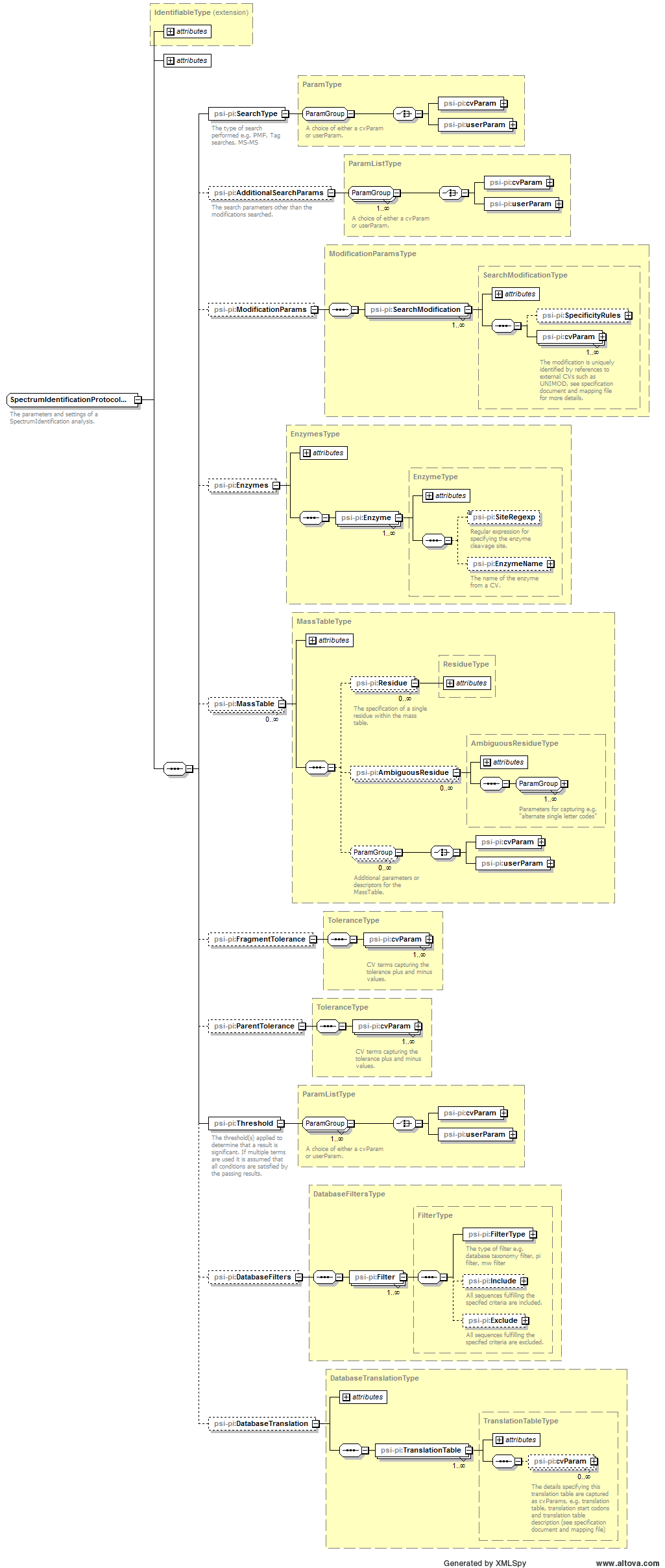
| Attribute Name | Data Type | Use | Definition |
|---|
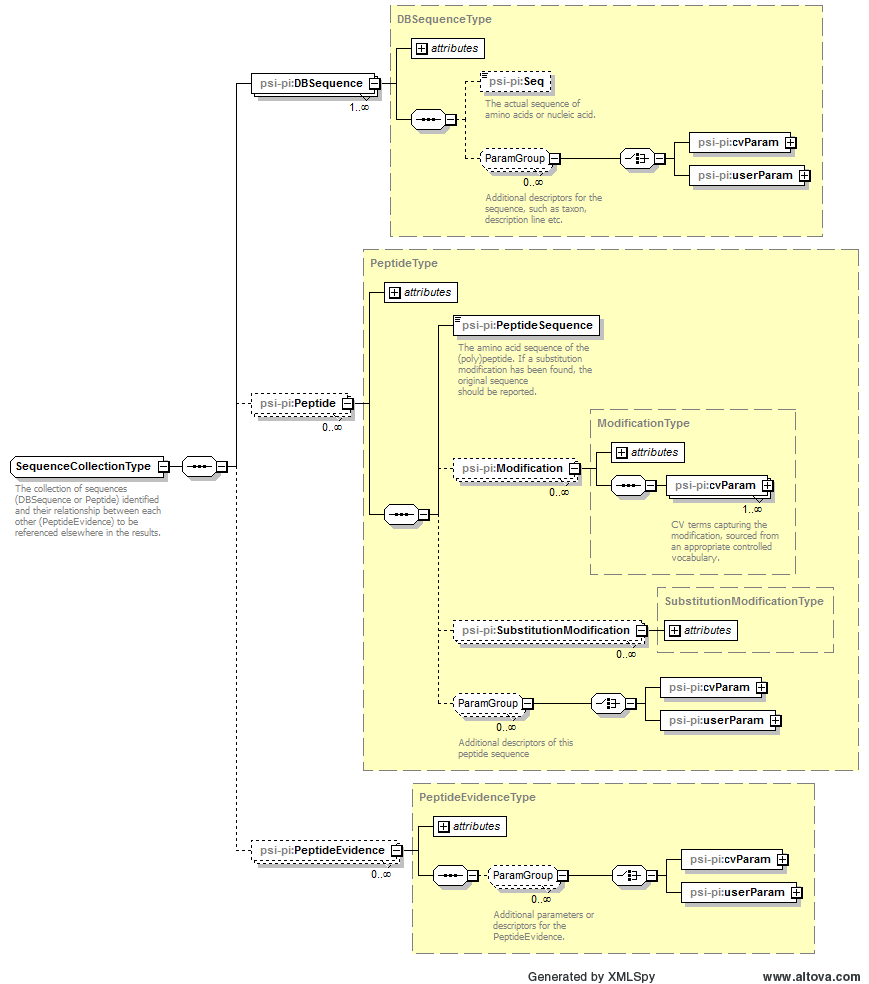
| dBSequence_ref | xsd:string | required | A reference to the protein sequence in which the specified peptide has been linked. |
| end | xsd:int | optional | The index position of the last amino acid of the peptide inside the protein sequence, where the first amino acid of the protein sequence is position 1. Must be provided unless this is a de novo search. |
| frame | allowed_frames | optional | The translation frame of this sequence if this is PeptideEvidence derived from nucleic acid sequence |
| id | xsd:string | required | An identifier is an unambiguous string that is unique within the scope (i.e. a document, a set of related documents, or a repository) of its use. |
| isDecoy | xsd:boolean | optional | Set to true if the peptide is matched to a decoy sequence. |
| name | xsd:string | optional | The potentially ambiguous common identifier, such as a human-readable name for the instance. |
| peptide_ref | xsd:string | required | A reference to the identified (poly)peptide sequence in the Peptide element. |
| post | xsd:string with restriction
[ABCDEFGHIJKLMNOPQRSTUVWXYZ?\-]{1} | optional | Post flanking residue. If the peptide is C-terminal, post="-" and not post="". If for any reason it is unknown (e.g. denovo), post="?" should be used. |
| pre | xsd:string with restriction
[ABCDEFGHIJKLMNOPQRSTUVWXYZ?\-]{1} | optional | Previous flanking residue. If the peptide is N-terminal, pre="-" and not pre="". If for any reason it is unknown (e.g. denovo), pre="?" should be used. |
| start | xsd:int | optional | Start position of the peptide inside the protein sequence, where the first amino acid of the protein sequence is position 1. Must be provided unless this is a de novo search. |
| translationTable_ref | xsd:string | optional | A reference to the translation table used if this is PeptideEvidence derived from nucleic acid sequence |