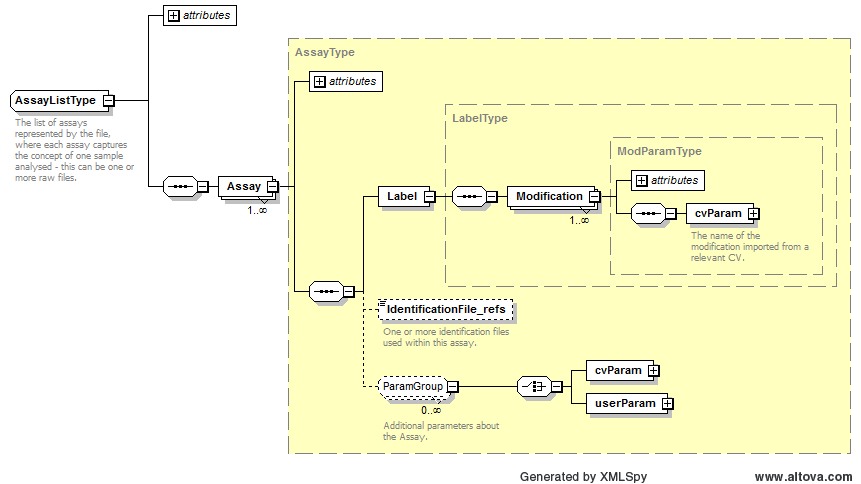
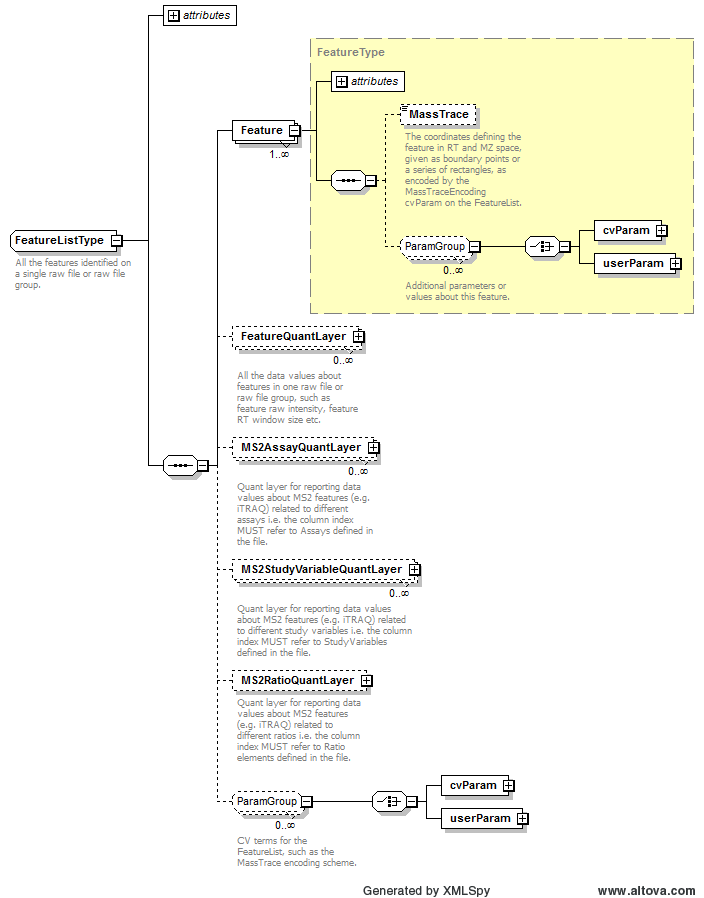
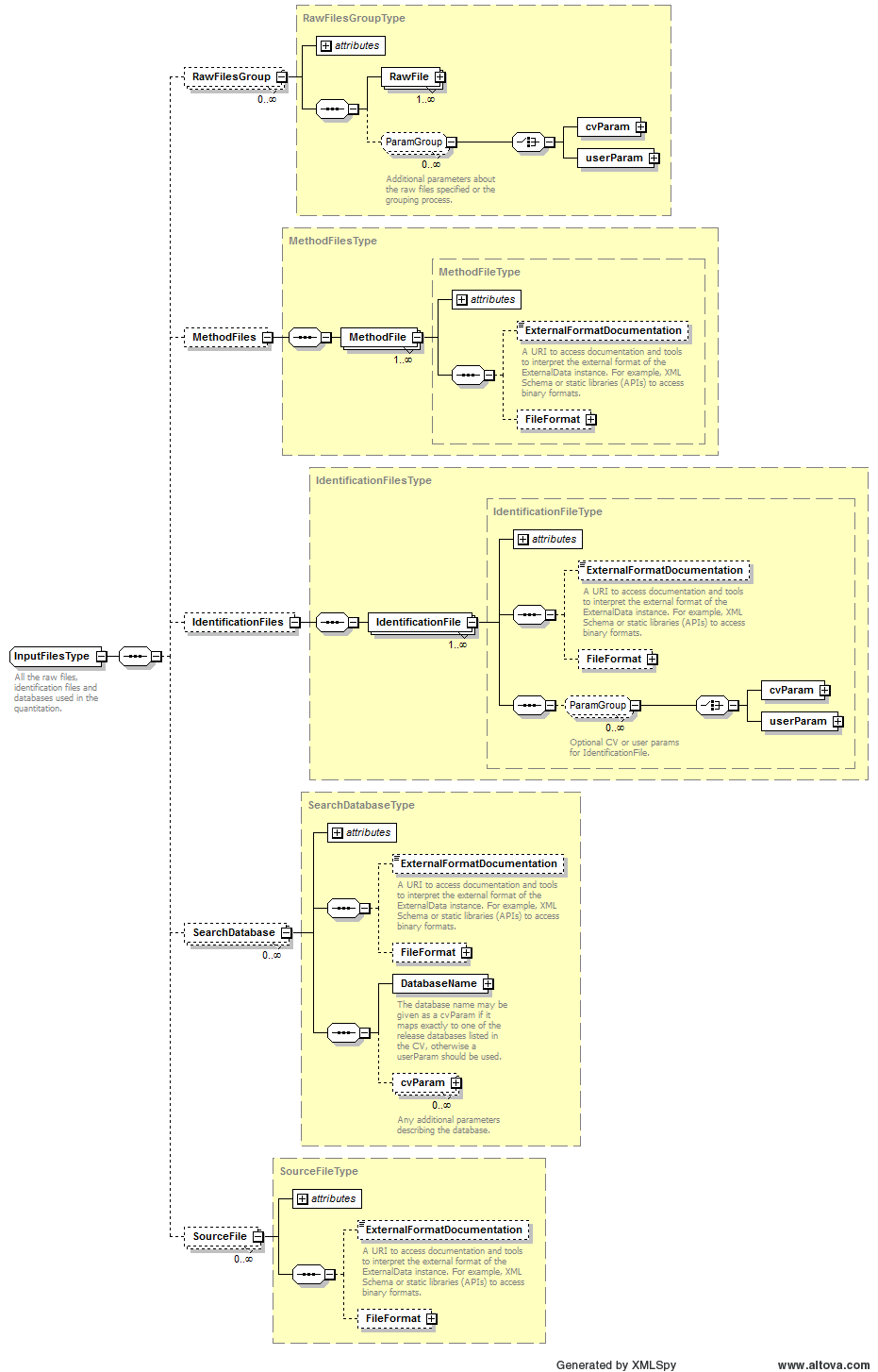
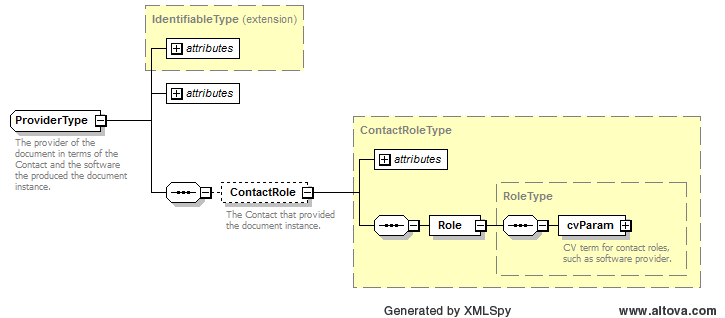
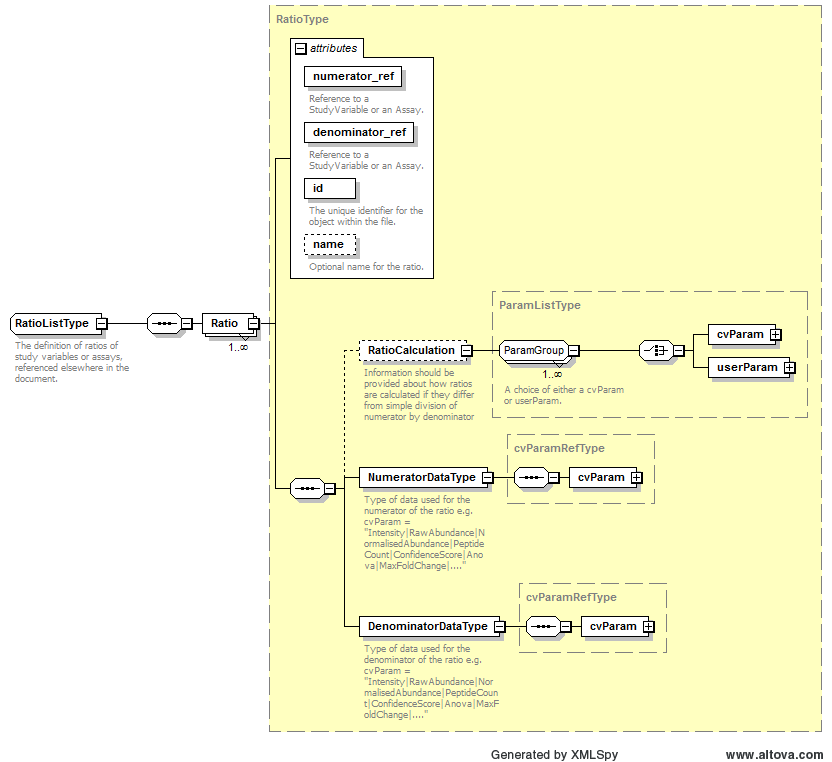
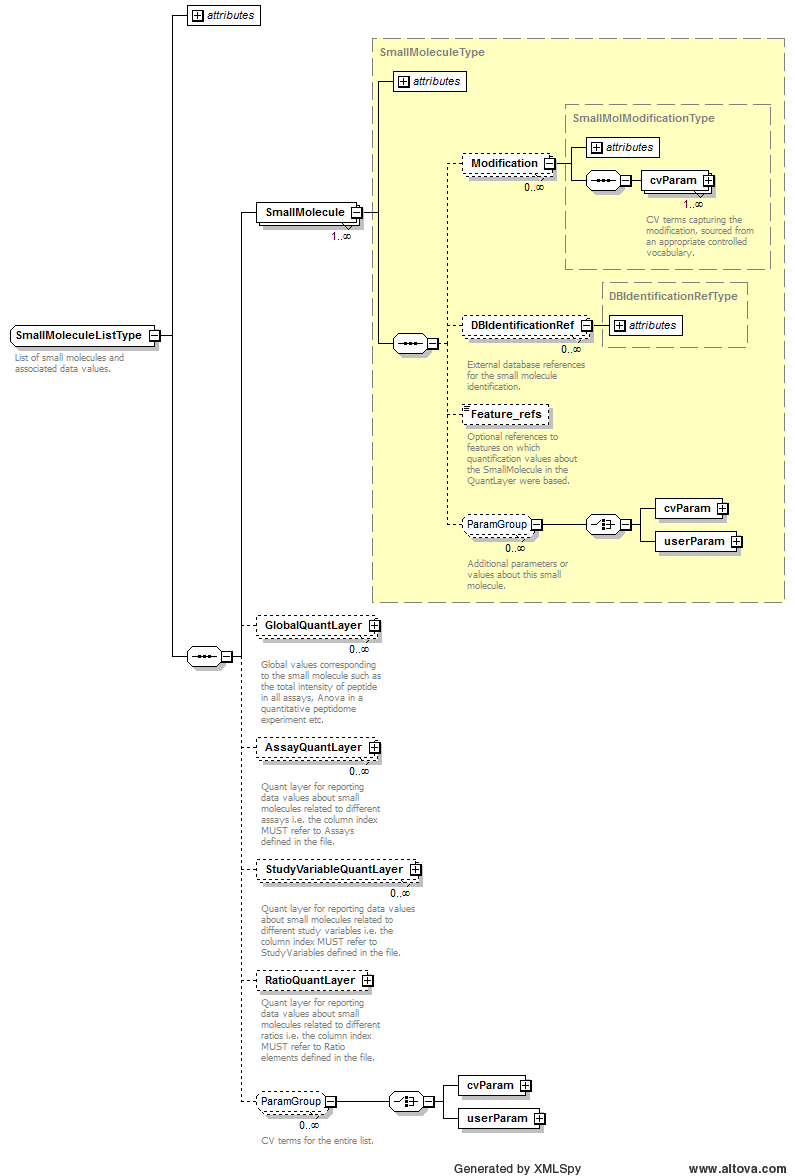
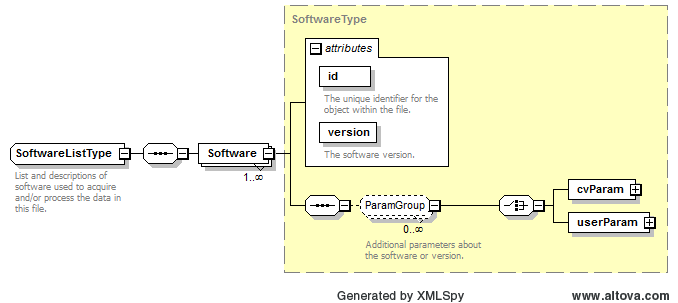
Path /MzQuantML/SmallMoleculeList/GlobalQuantLayer/ColumnDefinition/Column/DataType
MUST supply a *child* term of MS:1001805 () only once
e.g.: MS:1001130 ()
e.g.: MS:1001131 ()
e.g.: MS:1001132 ()
e.g.: MS:1001133 ()
e.g.: MS:1001134 ()
e.g.: MS:1001135 ()
e.g.: MS:1001136 ()
e.g.: MS:1001137 ()
e.g.: MS:1001138 ()
e.g.: MS:1001141 ()
et al.
MUST supply a *child* term of MS:1001405 () only once
e.g.: MS:1000796 ()
e.g.: MS:1000797 ()
e.g.: MS:1000798 ()
e.g.: MS:1000903 ()
e.g.: MS:1000904 ()
e.g.: MS:1000926 ()
e.g.: MS:1001030 ()
e.g.: MS:1001035 ()
e.g.: MS:1001036 ()
e.g.: MS:1001088 ()
et al.
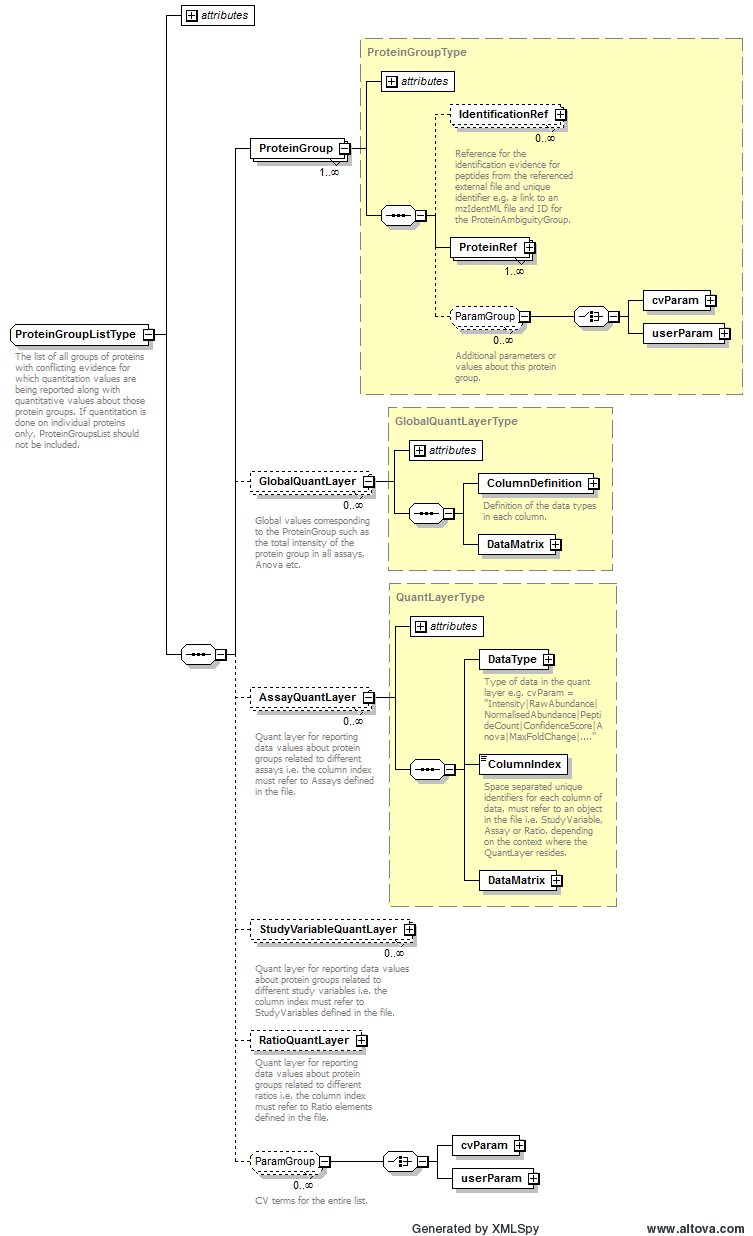
Path /MzQuantML/ProteinGroupList/AssayQuantLayer/DataType
MUST supply a *child* term of MS:1001805 () only once
e.g.: MS:1001130 ()
e.g.: MS:1001131 ()
e.g.: MS:1001132 ()
e.g.: MS:1001133 ()
e.g.: MS:1001134 ()
e.g.: MS:1001135 ()
e.g.: MS:1001136 ()
e.g.: MS:1001137 ()
e.g.: MS:1001138 ()
e.g.: MS:1001141 ()
et al.
MUST supply a *child* term of MS:1001405 () only once
e.g.: MS:1000796 ()
e.g.: MS:1000797 ()
e.g.: MS:1000798 ()
e.g.: MS:1000903 ()
e.g.: MS:1000904 ()
e.g.: MS:1000926 ()
e.g.: MS:1001030 ()
e.g.: MS:1001035 ()
e.g.: MS:1001036 ()
e.g.: MS:1001088 ()
et al.
Path /MzQuantML/ProteinGroupList/GlobalQuantLayer/ColumnDefinition/Column/DataType
MUST supply a *child* term of MS:1001805 () only once
e.g.: MS:1001130 ()
e.g.: MS:1001131 ()
e.g.: MS:1001132 ()
e.g.: MS:1001133 ()
e.g.: MS:1001134 ()
e.g.: MS:1001135 ()
e.g.: MS:1001136 ()
e.g.: MS:1001137 ()
e.g.: MS:1001138 ()
e.g.: MS:1001141 ()
et al.
MUST supply a *child* term of MS:1001405 () only once
e.g.: MS:1000796 ()
e.g.: MS:1000797 ()
e.g.: MS:1000798 ()
e.g.: MS:1000903 ()
e.g.: MS:1000904 ()
e.g.: MS:1000926 ()
e.g.: MS:1001030 ()
e.g.: MS:1001035 ()
e.g.: MS:1001036 ()
e.g.: MS:1001088 ()
et al.
Path /MzQuantML/FeatureList/MS2AssayQuantLayer/DataType
MUST supply a *child* term of MS:1001805 () only once
e.g.: MS:1001130 ()
e.g.: MS:1001131 ()
e.g.: MS:1001132 ()
e.g.: MS:1001133 ()
e.g.: MS:1001134 ()
e.g.: MS:1001135 ()
e.g.: MS:1001136 ()
e.g.: MS:1001137 ()
e.g.: MS:1001138 ()
e.g.: MS:1001141 ()
et al.
MUST supply a *child* term of MS:1001405 () only once
e.g.: MS:1000796 ()
e.g.: MS:1000797 ()
e.g.: MS:1000798 ()
e.g.: MS:1000903 ()
e.g.: MS:1000904 ()
e.g.: MS:1000926 ()
e.g.: MS:1001030 ()
e.g.: MS:1001035 ()
e.g.: MS:1001036 ()
e.g.: MS:1001088 ()
et al.
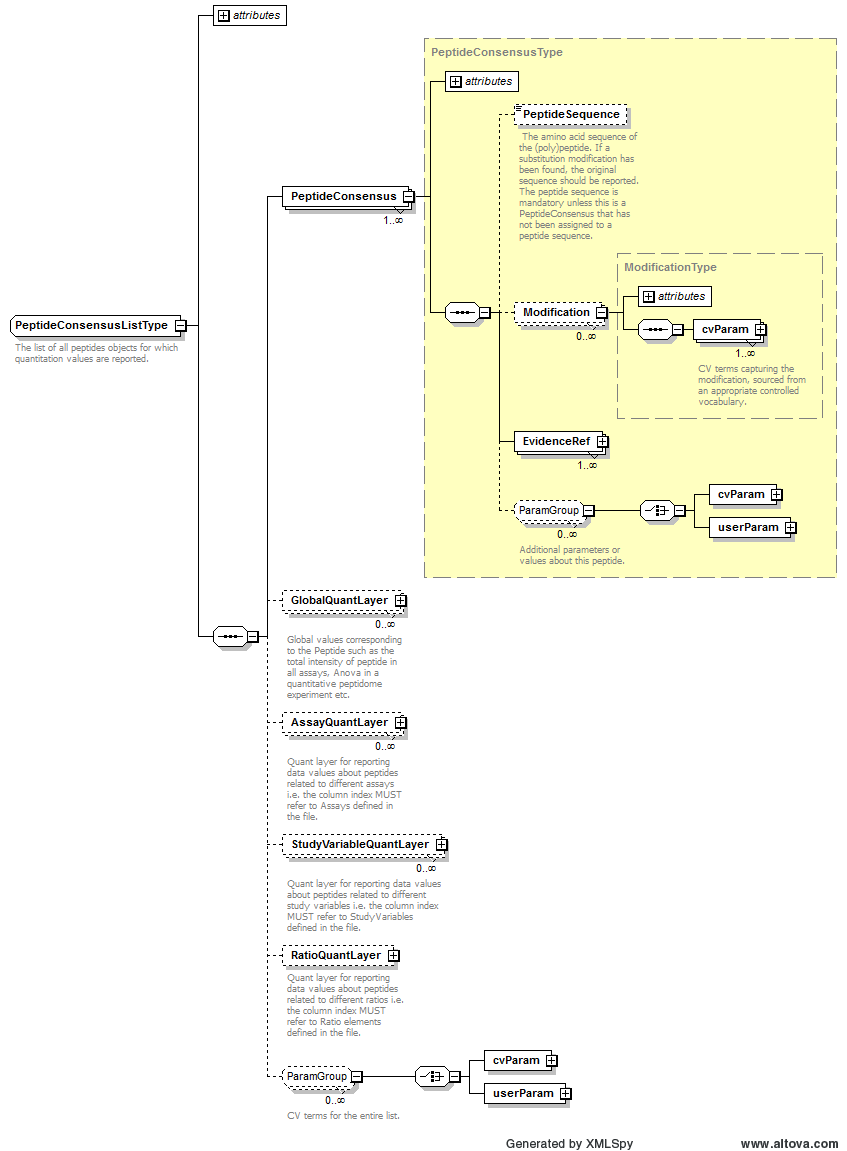
Path /MzQuantML/PeptideConsensusList/AssayQuantLayer/DataType
MUST supply a *child* term of MS:1001805 () only once
e.g.: MS:1001130 ()
e.g.: MS:1001131 ()
e.g.: MS:1001132 ()
e.g.: MS:1001133 ()
e.g.: MS:1001134 ()
e.g.: MS:1001135 ()
e.g.: MS:1001136 ()
e.g.: MS:1001137 ()
e.g.: MS:1001138 ()
e.g.: MS:1001141 ()
et al.
MUST supply a *child* term of MS:1001405 () only once
e.g.: MS:1000796 ()
e.g.: MS:1000797 ()
e.g.: MS:1000798 ()
e.g.: MS:1000903 ()
e.g.: MS:1000904 ()
e.g.: MS:1000926 ()
e.g.: MS:1001030 ()
e.g.: MS:1001035 ()
e.g.: MS:1001036 ()
e.g.: MS:1001088 ()
et al.
Path /MzQuantML/ProteinGroupList/StudyVariableQuantLayer/DataType
MUST supply a *child* term of MS:1001805 () only once
e.g.: MS:1001130 ()
e.g.: MS:1001131 ()
e.g.: MS:1001132 ()
e.g.: MS:1001133 ()
e.g.: MS:1001134 ()
e.g.: MS:1001135 ()
e.g.: MS:1001136 ()
e.g.: MS:1001137 ()
e.g.: MS:1001138 ()
e.g.: MS:1001141 ()
et al.
MUST supply a *child* term of MS:1001405 () only once
e.g.: MS:1000796 ()
e.g.: MS:1000797 ()
e.g.: MS:1000798 ()
e.g.: MS:1000903 ()
e.g.: MS:1000904 ()
e.g.: MS:1000926 ()
e.g.: MS:1001030 ()
e.g.: MS:1001035 ()
e.g.: MS:1001036 ()
e.g.: MS:1001088 ()
et al.
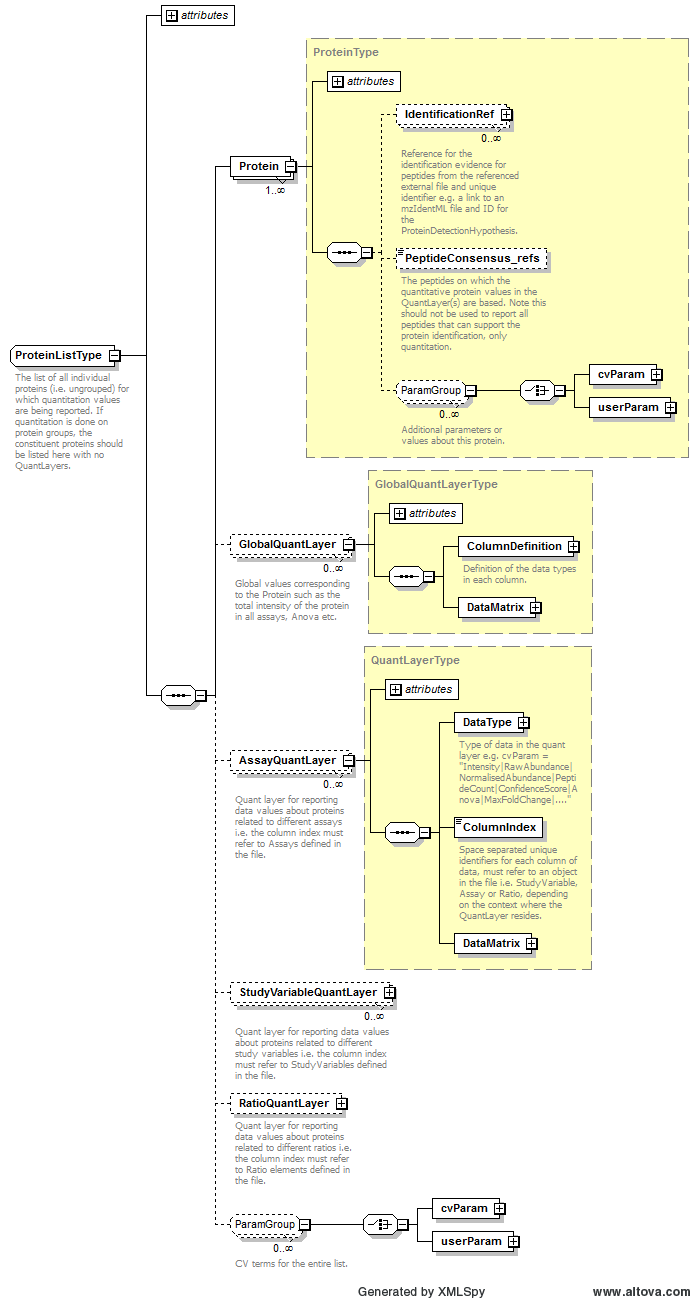
Path /MzQuantML/ProteinList/GlobalQuantLayer/ColumnDefinition/Column/DataType
MUST supply a *child* term of MS:1001805 () only once
e.g.: MS:1001130 ()
e.g.: MS:1001131 ()
e.g.: MS:1001132 ()
e.g.: MS:1001133 ()
e.g.: MS:1001134 ()
e.g.: MS:1001135 ()
e.g.: MS:1001136 ()
e.g.: MS:1001137 ()
e.g.: MS:1001138 ()
e.g.: MS:1001141 ()
et al.
MUST supply a *child* term of MS:1001405 () only once
e.g.: MS:1000796 ()
e.g.: MS:1000797 ()
e.g.: MS:1000798 ()
e.g.: MS:1000903 ()
e.g.: MS:1000904 ()
e.g.: MS:1000926 ()
e.g.: MS:1001030 ()
e.g.: MS:1001035 ()
e.g.: MS:1001036 ()
e.g.: MS:1001088 ()
et al.
Path /MzQuantML/SmallMoleculeList/StudyVariableQuantLayer/DataType
MUST supply a *child* term of MS:1001805 () only once
e.g.: MS:1001130 ()
e.g.: MS:1001131 ()
e.g.: MS:1001132 ()
e.g.: MS:1001133 ()
e.g.: MS:1001134 ()
e.g.: MS:1001135 ()
e.g.: MS:1001136 ()
e.g.: MS:1001137 ()
e.g.: MS:1001138 ()
e.g.: MS:1001141 ()
et al.
MUST supply a *child* term of MS:1001405 () only once
e.g.: MS:1000796 ()
e.g.: MS:1000797 ()
e.g.: MS:1000798 ()
e.g.: MS:1000903 ()
e.g.: MS:1000904 ()
e.g.: MS:1000926 ()
e.g.: MS:1001030 ()
e.g.: MS:1001035 ()
e.g.: MS:1001036 ()
e.g.: MS:1001088 ()
et al.
Path /MzQuantML/FeatureList/FeatureQuantLayer/ColumnDefinition/Column/DataType
MUST supply a *child* term of MS:1001805 () only once
e.g.: MS:1001130 ()
e.g.: MS:1001131 ()
e.g.: MS:1001132 ()
e.g.: MS:1001133 ()
e.g.: MS:1001134 ()
e.g.: MS:1001135 ()
e.g.: MS:1001136 ()
e.g.: MS:1001137 ()
e.g.: MS:1001138 ()
e.g.: MS:1001141 ()
et al.
MUST supply a *child* term of MS:1001405 () only once
e.g.: MS:1000796 ()
e.g.: MS:1000797 ()
e.g.: MS:1000798 ()
e.g.: MS:1000903 ()
e.g.: MS:1000904 ()
e.g.: MS:1000926 ()
e.g.: MS:1001030 ()
e.g.: MS:1001035 ()
e.g.: MS:1001036 ()
e.g.: MS:1001088 ()
et al.
Path /MzQuantML/FeatureList/MS2StudyVariableQuantLayer/DataType
MUST supply a *child* term of MS:1001805 () only once
e.g.: MS:1001130 ()
e.g.: MS:1001131 ()
e.g.: MS:1001132 ()
e.g.: MS:1001133 ()
e.g.: MS:1001134 ()
e.g.: MS:1001135 ()
e.g.: MS:1001136 ()
e.g.: MS:1001137 ()
e.g.: MS:1001138 ()
e.g.: MS:1001141 ()
et al.
MUST supply a *child* term of MS:1001405 () only once
e.g.: MS:1000796 ()
e.g.: MS:1000797 ()
e.g.: MS:1000798 ()
e.g.: MS:1000903 ()
e.g.: MS:1000904 ()
e.g.: MS:1000926 ()
e.g.: MS:1001030 ()
e.g.: MS:1001035 ()
e.g.: MS:1001036 ()
e.g.: MS:1001088 ()
et al.
Path /MzQuantML/ProteinList/StudyVariableQuantLayer/DataType
MUST supply a *child* term of MS:1001805 () only once
e.g.: MS:1001130 ()
e.g.: MS:1001131 ()
e.g.: MS:1001132 ()
e.g.: MS:1001133 ()
e.g.: MS:1001134 ()
e.g.: MS:1001135 ()
e.g.: MS:1001136 ()
e.g.: MS:1001137 ()
e.g.: MS:1001138 ()
e.g.: MS:1001141 ()
et al.
MUST supply a *child* term of MS:1001405 () only once
e.g.: MS:1000796 ()
e.g.: MS:1000797 ()
e.g.: MS:1000798 ()
e.g.: MS:1000903 ()
e.g.: MS:1000904 ()
e.g.: MS:1000926 ()
e.g.: MS:1001030 ()
e.g.: MS:1001035 ()
e.g.: MS:1001036 ()
e.g.: MS:1001088 ()
et al.
Path /MzQuantML/SmallMoleculeList/AssayQuantLayer/DataType
MUST supply a *child* term of MS:1001805 () only once
e.g.: MS:1001130 ()
e.g.: MS:1001131 ()
e.g.: MS:1001132 ()
e.g.: MS:1001133 ()
e.g.: MS:1001134 ()
e.g.: MS:1001135 ()
e.g.: MS:1001136 ()
e.g.: MS:1001137 ()
e.g.: MS:1001138 ()
e.g.: MS:1001141 ()
et al.
MUST supply a *child* term of MS:1001405 () only once
e.g.: MS:1000796 ()
e.g.: MS:1000797 ()
e.g.: MS:1000798 ()
e.g.: MS:1000903 ()
e.g.: MS:1000904 ()
e.g.: MS:1000926 ()
e.g.: MS:1001030 ()
e.g.: MS:1001035 ()
e.g.: MS:1001036 ()
e.g.: MS:1001088 ()
et al.
Path /MzQuantML/PeptideConsensusList/StudyVariableQuantLayer/DataType
MUST supply a *child* term of MS:1001805 () only once
e.g.: MS:1001130 ()
e.g.: MS:1001131 ()
e.g.: MS:1001132 ()
e.g.: MS:1001133 ()
e.g.: MS:1001134 ()
e.g.: MS:1001135 ()
e.g.: MS:1001136 ()
e.g.: MS:1001137 ()
e.g.: MS:1001138 ()
e.g.: MS:1001141 ()
et al.
MUST supply a *child* term of MS:1001405 () only once
e.g.: MS:1000796 ()
e.g.: MS:1000797 ()
e.g.: MS:1000798 ()
e.g.: MS:1000903 ()
e.g.: MS:1000904 ()
e.g.: MS:1000926 ()
e.g.: MS:1001030 ()
e.g.: MS:1001035 ()
e.g.: MS:1001036 ()
e.g.: MS:1001088 ()
et al.
Path /MzQuantML/ProteinList/AssayQuantLayer/DataType
MUST supply a *child* term of MS:1001805 () only once
e.g.: MS:1001130 ()
e.g.: MS:1001131 ()
e.g.: MS:1001132 ()
e.g.: MS:1001133 ()
e.g.: MS:1001134 ()
e.g.: MS:1001135 ()
e.g.: MS:1001136 ()
e.g.: MS:1001137 ()
e.g.: MS:1001138 ()
e.g.: MS:1001141 ()
et al.
MUST supply a *child* term of MS:1001405 () only once
e.g.: MS:1000796 ()
e.g.: MS:1000797 ()
e.g.: MS:1000798 ()
e.g.: MS:1000903 ()
e.g.: MS:1000904 ()
e.g.: MS:1000926 ()
e.g.: MS:1001030 ()
e.g.: MS:1001035 ()
e.g.: MS:1001036 ()
e.g.: MS:1001088 ()
et al.
Path /MzQuantML/PeptideConsensusList/GlobalQuantLayer/ColumnDefinition/Column/DataType
MUST supply a *child* term of MS:1001805 () only once
e.g.: MS:1001130 ()
e.g.: MS:1001131 ()
e.g.: MS:1001132 ()
e.g.: MS:1001133 ()
e.g.: MS:1001134 ()
e.g.: MS:1001135 ()
e.g.: MS:1001136 ()
e.g.: MS:1001137 ()
e.g.: MS:1001138 ()
e.g.: MS:1001141 ()
et al.
MUST supply a *child* term of MS:1001405 () only once
e.g.: MS:1000796 ()
e.g.: MS:1000797 ()
e.g.: MS:1000798 ()
e.g.: MS:1000903 ()
e.g.: MS:1000904 ()
e.g.: MS:1000926 ()
e.g.: MS:1001030 ()
e.g.: MS:1001035 ()
e.g.: MS:1001036 ()
e.g.: MS:1001088 ()
et al.
|